Complex bacterial vaccines contain multiple components, including protein antigens, lipopolysaccharides, capsule and other macromolecules. Although they are known to provide effective protection against disease, the way in which multiple components combine to elicit a protective response is poorly understood. One way to obtain insight into this important question is by using dedicated protein antigen microarrays. Immobilised peptides or proteins on a microarray are exposed to serum from vaccinated individuals. Through high-resolution scanning, the profile of antibodies against each antigen can be accurately quantified.
We have demonstrated that machine learning methods can be effectively applied to extract valuable information on the role of individual antigens in stimulating humoral immune responses.
The combination of high-quality instrumentation for quantifying bound antibody, with protein science and machine learning methods, is a powerful approach that could revolutionise our design of complex vaccines.
Background
The design and formulation of complex vaccines against bacterial disease poses a significant challenge. Traditionally, such vaccines were generated by chemical extraction or the use of an attenuated strain. An example is outer membrane vesicles (OMVs), which are membrane-rich extracts isolated by detergent extraction from the bacterium. Although they can be effective in protecting against infection, this protection is limited to strains that are similar to the ‘parent’ used for OMV extraction. Manipulation of the OMV content, which is highly complex, is challenging: strains can be engineered to modify LPS and up- or down-regulate specific antigens, but controlling precise quantities is difficult. Purified recombinant proteins can be added to the OMV, in combination with a suitable adjuvant, such as the 4CMenB vaccine developed by GSKt for he treatment of meningococcal meningitis [1].
Although 4CMenB has been shown to be effective in reducing cases of serogroup B meningococcal meningitis, the precise combination of antigens within the vaccine which delivers protection is less clear.
Interest in this vaccine has increased recently, due to the observation that 4CMenB provides some protection, although limited, against gonorrhoea [2,3].
Both diseases are caused by related organisms: Neisseria meningitidis (meningitis) and Neisseria gonorrhoeae (gonorrhoea).
How is it that a vaccine which was designed to protect against one disease can be effective against another, and which components within 4CMenB might be responsible?
We set out to address these questions using designed microarrays, which incorporated major protein antigens from both organisms. At the heart of the approach is the use of the InnoScan 710 scanner to accurately quantify antibody levels against each antigen over a scale of at least two orders of magnitude. These datasets have revealed the complexity of the antibody responses to the 4CMenB and show how cross-reactivity against antigens from each organism can explain the protection observed in the clinic.
Principles and process: immunoprofiling using antigen microarray and InnoScan 710
Peptide and protein microarrays provide significantly more detailed information on polyclonal antibody specificities against multiple antigens compared to traditional methods, such as ELISA. The principle of the process is shown in Figure 1:
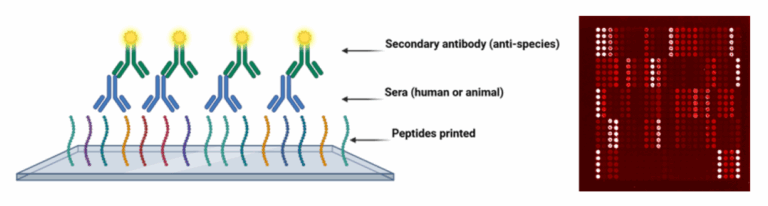
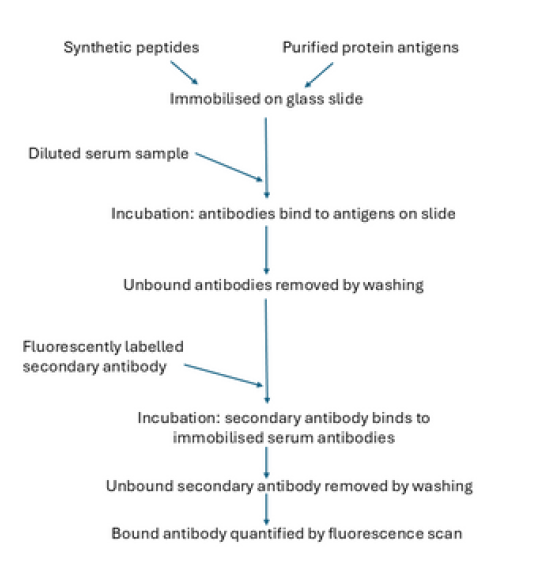
The general process is summarised in Figure 2. We use a range of in silico methods to select antigens for deposition into the array. For peptides, B- or T-cell epitope prediction methods are valuable in identifying regions of whole protein antigens which are most likely to be the target of antibody binding. Fabrication of whole protein arrays is a more complex process that involves the expression and purification of recombinant proteins. The selection of suitable antigens can utilise a variety of information sources, including proteomic datasets and genomic analysis. Control antibodies are included in each array, which allows for standardisation of reactivity against the secondary antibody.
We observe minor deviations from linearity of fluorescence against the quantity of deposited antibody, which can be readily corrected. Responses are sensitive over at least two orders of magnitude, which allows detection of low amounts of antibodies. Dilution of serum requires optimisation – optimal concentrations differ with source (mouse, human) and subclass (e.g., IgG1, IgG2a, etc). Once an optimal dilution is identified, it is applied to all samples.
Data are collected using the InnoScan 710 scanner, and the laser and photomultiplier (PMT) settings are determined based on the control spots included in the array.
Examples: microarrays for specific antigens
Example 1: A microarray specific for meningococcal meningitis antigens.
In this study, we used a designed microarray with about 90 different protein antigens from N. meningitidis, which we probed using sera from volunteers vaccinated with the 4CMenB contains an OMV component, and hence, many hundreds of potential protein antigens are present.
We collected antigen reactivity profiles for sera from individuals before they received their first dose and then after receiving two and three doses.
In Figure 3, each point is a serum sample, with the antigen-specific responses condensed to a single point in 2D space. Data points close in space in this plot imply similar antigen reactivity profiles. Hence, when the vaccine is administered, we observe a shift in sample responses, as evident from a comparison of the pre-vaccination data in purple with responses after a second dose (green) and a third dose (red). Using tools like this, we can track immune responses to complex, multicomponent vaccines, such as 4CMenB.
![Figure 3: ASCA separation of serum samples against a meningitis antigen panel microarray. The ellipses show the centres of each group. Data used are taken from Ramirez-Bencomo et al. [4]](https://www.innopsys.com/wp-content/uploads/2025/11/Capture-decran-2025-11-20-122955-768x429.png)
Example 2: A microarray specific for gonorrhoea antigens
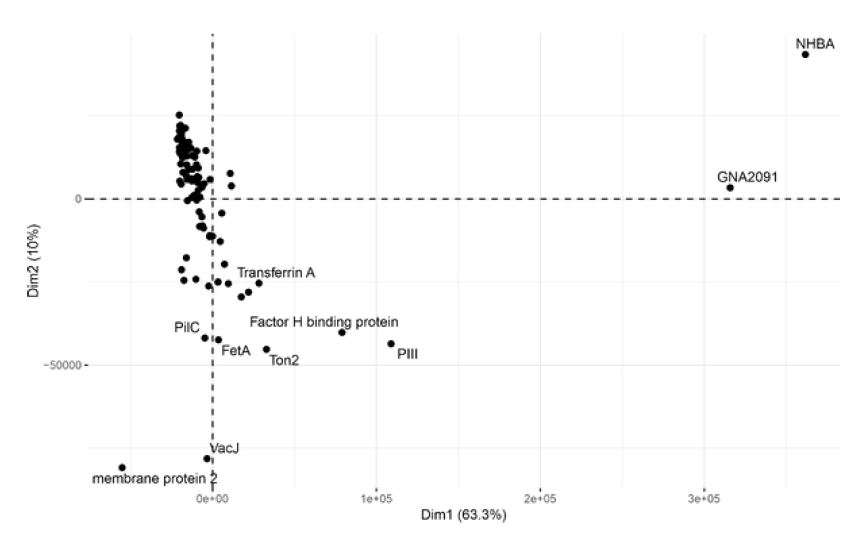
There is evidence that 4CMenB can also protect against infection by N. gonorrhoeae, the causative agent of gonorrhoea. Although N. meningitidis and N. gonorrhoeae are related bacteria, the sequences of their surface proteins are not identical. Using a dedicated microarray, we set out to examine whether vaccination with 4CMenB was able to induce antibodies which cross-react with gonorrhoea antigens [5]. This involved a similar experiment to Example 1, where sera from a cohort of volunteers were collected before and after vaccination. In Figure 4, we present an example of an alternative approach to analysing the data, focusing on the antigens responsible for the primary antibody response. In this figure, we chart the antigens that are principally responsible for stimulating IgG antibodies after vaccination. Using this kind of analysis, we can start to understand the potential basis for cross-protection of 4CMenB against gonorrhoea infection.
Conclusion: immunoprofiling using microarrays
These examples demonstrate the potential for utilising dedicated antigen microarrays to enhance understanding of immune responses to complex vaccines.
At the heart of implementing this technology is the use of the InnoScan 710 scanner, which enables rapid and reliable quantification of bound antibody to selected antigen panels.
These datasets can be analysed in multiple ways, exploiting the parallel development of machine learning methods, which can be applied to train them against metadata from vaccinated populations. For example, in the case of gonorrhoea, we can start to investigate whether there are differences in immune responses between men and women – a question of prime importance for a sexually transmitted disease.
We can also develop more sophisticated approaches to understand how the composition is linked to protection against infection, leading to the development of improved complex vaccines.
References
1] Bai, X., Findlow, J., & Borrow, R. (2011). Recombinant protein meningococcal serogroup B vaccine combined with outer membrane vesicles. Expert Opin Biol Ther, 11(7), 969-985. https://doi.org/10.1517/14712598.2011.585965
[2] Semchenko, E. A., Tan, A., Borrow, R., & Seib, K. L. (2019). The Serogroup B Meningococcal Vaccine Bexsero Elicits Antibodies to Neisseria gonorrhoeae. Clin Infect Dis, 69(7), 1101-1111.
https://doi.org/10.1093/cid/ciy1061
[3] Petousis-Harris, H., Paynter, J., Morgan, J., Saxton, P., McArdle, B., Goodyear-Smith, F., & Black, S. (2017). Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet, 390(10102), 1603-1610.
[4] Ramirez-Bencomo, F., A. Thistlethwaite, V. Viviani, E. Bartolini, M. Pizza, A. Biolchi, A. Muzzi, et al. “Identification of Immunogenic Outer Membrane Vesicle Vaccine Antigen Components Using a Meningococcal Protein Microarray.” [In eng]. Vaccine 53 (Apr 19 2025): 126953. https://doi.org/10.1016/j.vaccine.2025.126953. https://www.ncbi.nlm.nih.gov/pubmed/40043411
[5] Stejskal, L., A. Thistlethwaite, F. Ramirez-Bencomo, S. Rashmi, O. Harrison, I. M. Feavers, M. C. J. Maiden, et al. “Profiling Igg and Iga Antibody Responses during Vaccination and Infection in a High-Risk Gonorrhoea Population.” Nat Commun 15, no. 1 (Aug 7 2024): 6712. https://doi.org/10.1038/s41467-024-51053-x https://www.ncbi.nlm.nih.gov/pubmed/39112489
