Proteins are considered to be the key effectors of cellular response to external stimuli.
Differential expression of key proteins can modulate cell fate, either cell differentiation, proliferation or death. Protein arrays are a powerful tool for high-throughput screening of protein expression due to their capacity to miniaturize thousands of single points of analysis.
High-throughput screening requires of highly precise detection systems to increase assay sensitivity and dynamic range. The InnoScan microarray scanners allow for the detection of heterogeneous samples with weak and strong protein levels simultaneously.
Multiplexing is achieved through the detection of two (InnoScan 710 & 710-IR) or three (InnoScan 1100) fluorophores simultaneously, while the resolution and dynamic range are adapted for any type of protein array, either functional, analytical and reverse phase protein arrays.
This technical note is focused on the application of the InnoScan® 710 and 710IR microarray scanners to scan protein arrays. Its principal goal is to guide customers and distributors in choosing the adequate scanner according to the microarray they want to read. For that, a general explanation of protein array construction and applications is done and some tips to choose the more appropriate scanner are discussed.
What are protein arrays?
Protein arrays, also known as protein chips, are miniaturized, parallel assay systems that contain small amounts of either purified proteins or peptides in a high-density format. Protein array applications go from protein profiling to the discovering of new protein-protein, protein-DNA or protein-drug interactions.
The protein array experimental bases are the same as for DNA microarrays: purified proteins, peptides or cell extracts are immobilised on a solid support, usually modified glass slides resulting in arrays of proteins. In parallel, samples such as cell extracts, serum, tissue extracts or purified antibodies are labelled with a fluorescent tag molecule.
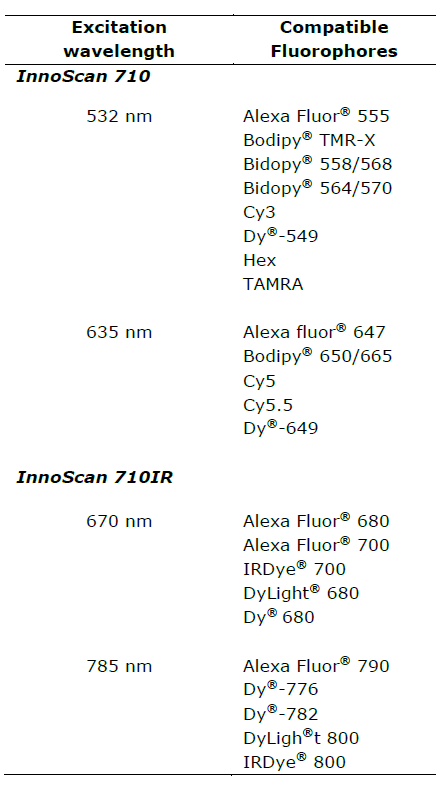
You can choose between the InnoScan 710 microarray scanner for detection of fluorophores on the classical 532nm and 635nm wavelengths or the InnoScan 710 IR microarray scanner for detection of the IR fluorescence at 670 nm and 785 nm of excitation wavelength.
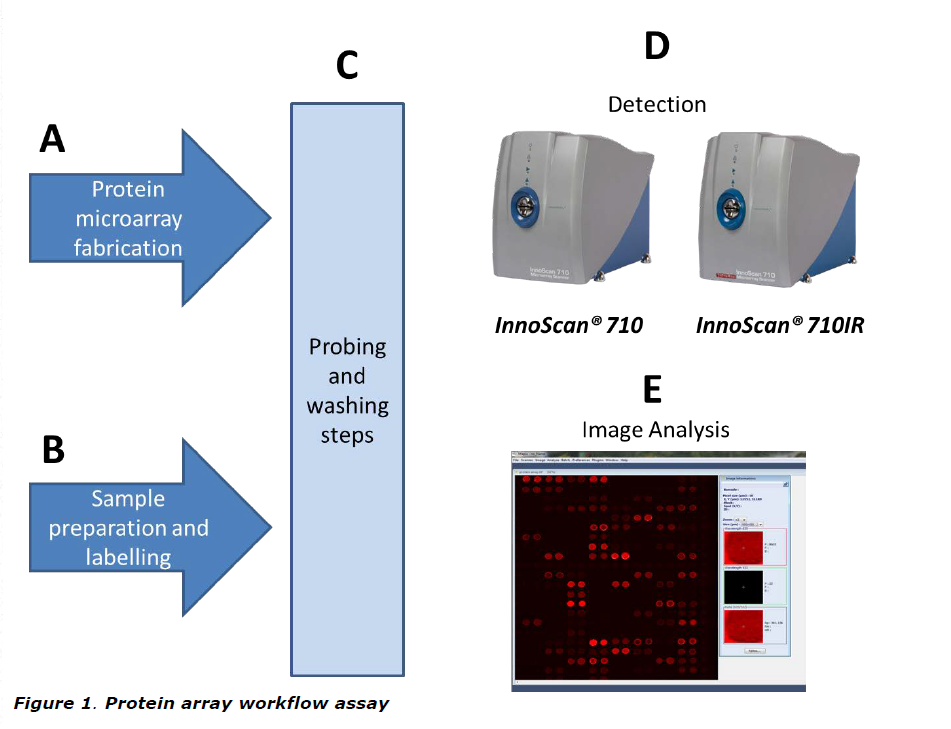
The resulting labelled samples are then probed with the immobilised proteins on the array. If targets of the immobilised proteins are present in the labelled samples, a fluorescent signal will be detected on the array (Figure 1).
Using protein arrays, researchers can evaluate the expression and/or the activity of hundreds or thousands of proteins in a single experiment. That is why the applications of protein arrays on basic and clinical research are very large. Proteome profiling could be now possible using proteome microarrays currently existing in the market.

Which are the different kinds of protein arrays?
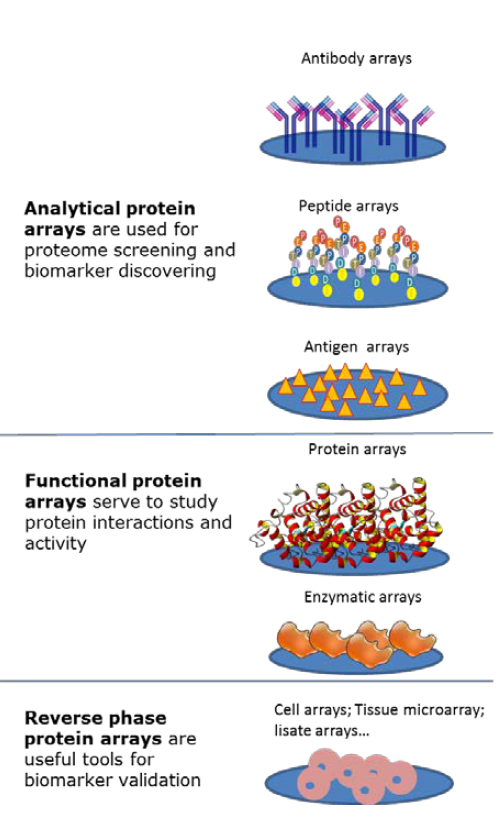
Protein arrays are classified in three groups according to the type of proteins that are immobilised on the slides and therefore their applications: analytical microarrays, functional arrays, and reverse phase arrays RPPA (Figure 2).
In general, analytical protein microarrays are constructed by arraying antibodies, purified proteins, well characterized peptides or non- folded antigens on the glass slide, which is then probed with a protein solution. This technique is based on the sandwich ELISA methodology and is used to measure protein expression levels in a solution or a sample.
Functional protein arrays are made of full-length functional proteins or protein domains attached to a functionalized matrix usually made of a layer of a polymer that confers a porous structure. They are used for studying protein interactions with other proteins, macromolecules, or small molecules such as ligand- receptor interactions, and for testing protein activity as it is the case of kinase arrays.
Reverse phase protein arrays (RPPA) correspond to the miniaturization of dot blotting. In RPPA, samples such cell lysates or plasma are spotted onto a glass slide using a robotic protein microarrayer; the arrayed samples are tested by using labeled antibodies against the protein of interest. RPPA is a useful tool for the evaluation of protein expression levels in hundreds of different samples in a single experiment.
Which are the applications of protein arrays?
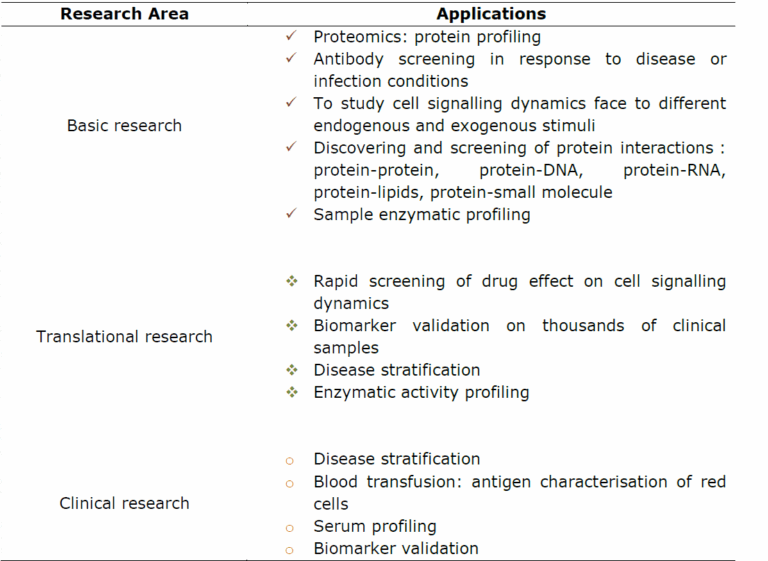
As mentioned above, the applications of protein arrays are very large. Table 1 summarizes some of them. Analytical protein arrays are mainly used to establish the protein expression profiles on complex samples. In this way, researchers can profile cellular responses at the protein level face to different stimuli such as environmental stress, diseases, infection or inflammation.
With functional protein arrays, protein interactions such as protein- protein, protein-DNA, protein-RNA, protein-phospholipid, and protein- small molecule interactions can be studied. Therefore, functional protein arrays are very useful in basic research to study the biochemical activities of several proteins in a single experiment.
RPPA arrays are potential tools for translational medicine and clinical research as they can be used for disease biomarker validation and disease stratification. For example, by monitoring biomarker dynamics in response to various doses of drugs at different disease stages, researchers can associate drug efficiency at each disease stage and then defining the best therapy for each disease stage which is one of the principal goals on personalized medicine.
Advantages of protein arrays against classical tests for protein expression and activity
Traditional approaches for protein research allows only for one or few proteins to be studied at a time. Such as time-intensive and money- consuming assays should not be suitable for systems biology and personalized medicine purposes. Proteomics based on two dimension electrophoresis combined with mass spectrometry is laborious and time- consuming. Furthermore, as part of this methodology; proteins have to be denatured which counteracts with protein-activity testing. Protein microarrays arise then as promising tools for elucidating the functional proteome by arraying intact proteins on glass slides coated with appropriate chemical treatments.
Why do several protein array protocols use IR labels?
One of the principal challenges on protein array technologies is to keep protein activity intact while immobilising proteins from complex samples. Modifying the chemistry of array’s supports, it is now possible to keep appropriate protein structure and activity for protein interactions assays. Advances in surface chemistry approaches have led to the development of different three dimensional supports that fit the requirements for protein stability.
Unfortunately, some of these supports give strong fluorescence signals when being read with classical microarray scanner wavelengths. This is especially true for nitrocellulose-coated glass slides which are widely used for the fabrication of protein arrays. The strong fluorescence produced by nitrocellulose translates into mediocre signal to noise ratio values, and therefore low sensitivity.
Using near-infrared (NIR) detection allows a dramatic decrease in background issues created by nitrocellulose and other supports. Samples are labelled with a NIR fluorophore to allow the target’s detection. NIR detection is known to increase SNR values and highly improve sensitivity for protein interaction assays.
How to choose the more appropriate scanner version?
Scanner selection depends on the fluorophores used for labelling. Not all protein array protocols are designed to be read on the NIR wavelengths. Table 2 lists fluorophores compatible with the different versions of the InnoScan 710 microarray scanner.
In general, protein arrays are made of spots of sizes of more than 50 microns of diameter for which the resolution given by the InnoScan 710 scanner is largely sufficient. For high density protein arrays with spot size less than 40µm the high image resolution given by the InnoScan 900 scanner is necessary.
Concluding remarks: protein microarrays
Advances in protein array technology have permitted a rapidly developing protein market. Protein array applications on disease biomarker discovery and validation reflect the enormous potential of protein arrays in the field of personalized medicine.
In harmony with the development of new protein array technologies, Innoscan 710 and 710IR microarray scanners deliver the high resolution and sensitivity required for this type of assay. Using the InnoScan 710IR microarrray scanner in combination with the appropriate infrared fluorophores, it is possible to avoid background fluorescence from supports such as nitrocellulose and thus increasing the sensitivity which is one of the principal challenges of protein array assays□
Disclaimers
InnoScan® is a registered trademark of INNOPSYS
Alexa Fluor® is a registered trademark of Molecular Probes (Life Technologies) Bidopy® is a registered trademark of Molecular Probes (Life Technologies)
Cy-Dye is a registered trademark of Amersham Pharmacia Biotech UK Ltd. Dy® is a trademark of Dynomics
DyLight® is a trademark of Thermo Fisher Scientific Inc. and its subsidiaries IRDye® is a registered trademark of Li-COR Biosciences