InnoScan microarray scanner: new publication
New Ultrabright RNA-Activated Fluorophore Developed Using Fragment-Based Screening
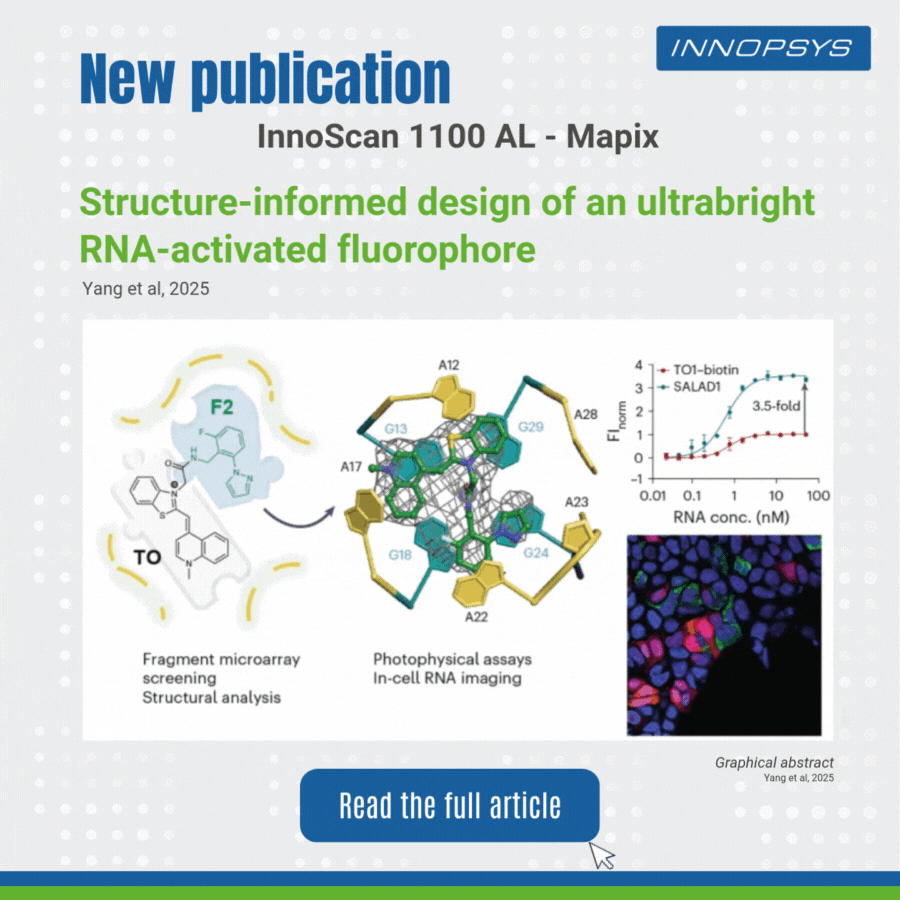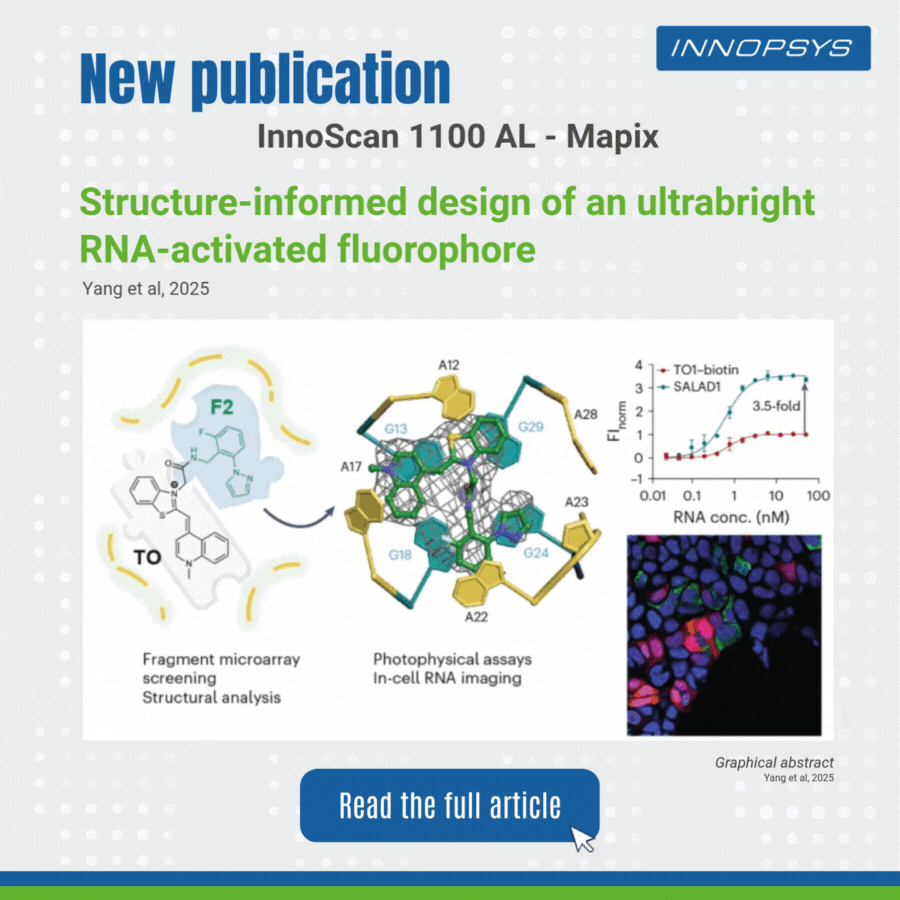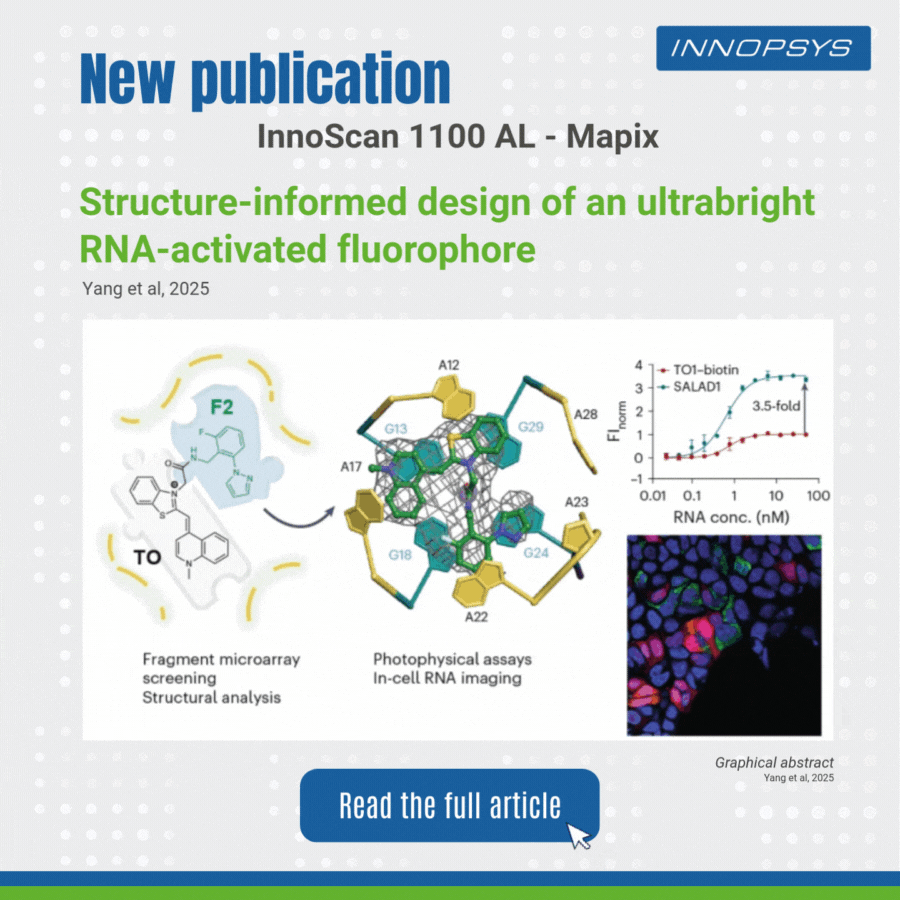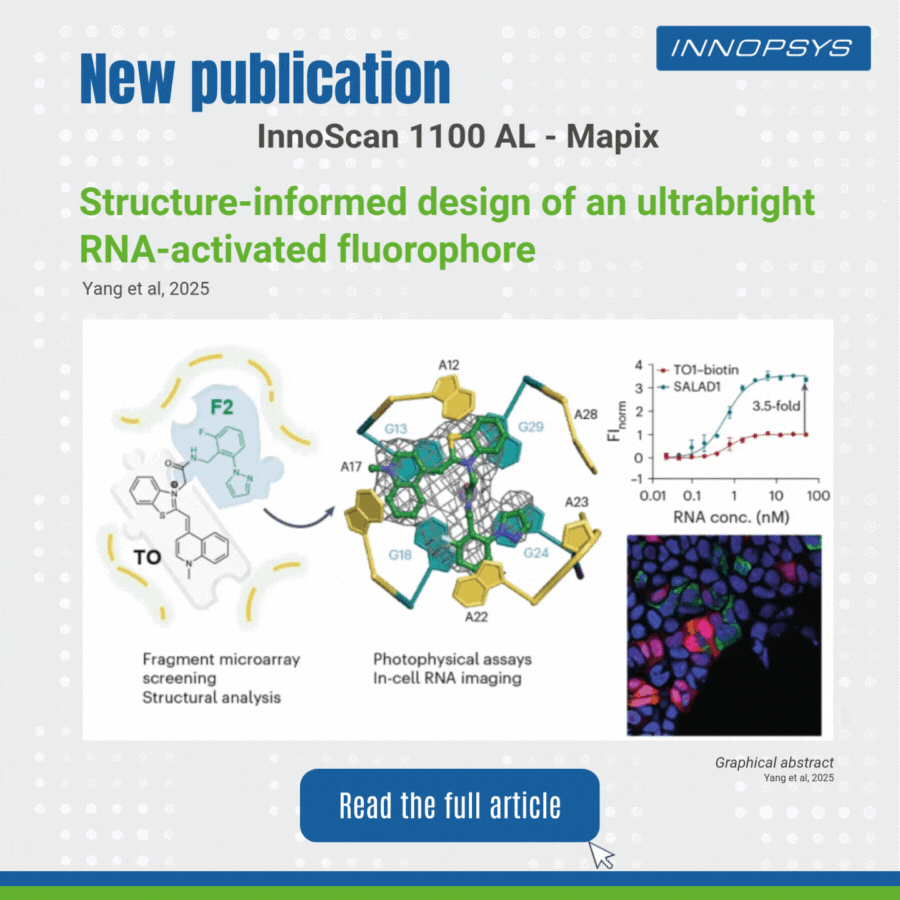
User case of the InnoScan Microarray scanner: new fluorophore development. In a recent study published in Nature Chemistry, researchers designed an ultrabright RNA-activated fluorophore, SALAD1, for advanced cellular RNA imaging. SALAD1 is a covalent hybrid created by linking TO (thiazole orange) and F2 fragment via an amide linker. This structure combines TO’s fluorogenic core with F2’s binding-enhancing properties, resulting in 3.5× brighter fluorescence than TO1-biotin.
The team employed a fragment-based small-molecule microarray (SMM) approach, leveraging the innoScan 1100 AL fluorescence scanner to screen 2,214 fragments for binding to the Mango II RNA aptamer. The innoScan system enabled high-throughput quantification of fluorescence intensities, identifying non-competitive fragments including F2 fragment which enhanced the brightness of thiazole orange (TO).
This work highlights how fragment-based design, aided by high-throughput tools like innoScan, can optimize RNA-targeting ligands for superior imaging.
Keywords: RNA imaging, fluorogenic aptamer, fragment-based screening, Mango II, InnoScan Microarray scanner: new fluorophore.
Original publication: Structure-informed design of an ultrabright RNA-activated fluorophore | Nature Chemistry
Learn more about the InnoScan series: InnoScan Microarray Scanners – Innopsys